PCOSBase Database
PCOSBase Schema
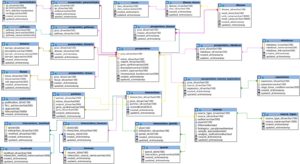
PCOSBase currently consists of 29 tables. Figure 1 shows all the 29 tables with the connections between table to table.
Figure 1: PCOSBase schema
Table Information
- pcosproteins. This is the main table of PCOSBase. This table has basic information including Entrez Gene ID, UniProt ID, gene symbol, gene description and the chromosomal location of all 8,185 PCOS-related proteins.
- geneontologies. This table contains the information of 13,237 gene ontologies that are associated with all PCOS-related proteins. GO ID, GO term, the definition/description of the GO term and ontology (BP, MF, CC) are provided.
- pcosproteins_geneontologies. This is the pivot table between pcosproteins and geneontologies The evidence is the source of GO annotation for particular PCOS-related proteins.
- pathways. This table lists 1,004 pathways obtained from three pathway databases, KEGG, BioCarta and WikiPathways.
- pcosproteins_pathways. This table links pcosproteins and pathways tables.
- domains. A total of 7,936 domains/families from InterPro database are listed in this table.
- crossrefs_domains. This table records all the signatures that contribute to domain entry. One domain id could have more than one signature. All of these data were retrieved from InterPro database.
- pcosproteins_domains. It links pcosproteins with domains tables.
- tissues. This table contains information of all the tissues and the cell types on where the PCOS-related proteins were expressed.
- pcosproteins_tissues. This table is the connective for pcosproteins and tissues tables.
- diseases. This table lists 1,928 diseases in Unified Medical Language System (UMLS) ID. Medical Subject Headings (MeSH) IDs are also provided in this table.
- classes. There are 39 classes of disease listed in this table.
- diseases_classes. This table classifies all the diseases in ‘diseases’ table into classes in ‘classes’ table. The diseases were classified based on MeSH database.
- pcosproteins_diseases. This table connects pcosproteins and diseases tables.
- partners. This table lists 16,847 proteins that interact with PCOS-related proteins.
- interactions. This table records all the interactions between PCOS-related proteins with the proteins in ‘partners’ table.
- imethods. This table lists 122 methods that have been used to detect the protein-protein interactions (PPI).
- interactions_imethods. This table links imethods with interactions table. It records which interactions have been detected using which methods.
- ipmids. This table contains 36,110 references from PubMed database for every interaction.
- interactions_ipmids. This table links interactions and ipmids.
- isources. This table has 24 sources (PPI databases and PPI publications), where all the interactions have been retrieved.
- interactions_isources. This table links which interactions were retrieved from.
- rdatabases. This table lists nine databases, where PCOS-related proteins have been retrieved.
- pcosproteins_rdatabases. This table links pcosproteins and rdatabases tables.
- source_types. This table contains only two source types which are transcriptomics and proteomics.
- sources. This table lists all the sources of PCOS-related proteins have been retrieved. All the sources are either gene expression studies (transcriptomics) or protein expression studies (proteomics).
- expressions. This table contains 28 expression patterns for the proteins in the sources tables.
- pcosproteins_sources_expressions. This table is a connective table between pcosproteins, sources and expressions tables.
- publications. This table is not connected to other tables. It listed all the publications that are related to PCOS.