Frequently asked questions (FAQs)
1. What is PCOSBase?
PCOSBase is a database that compiles all the proteins that are related to PCOS from differently publicly available databases and published expression studies, as listed in the PCOS-Related Proteins Data. PCOSBase also supplies additional information such as GO, pathways, domains, interactions, tissues and diseases that are associated with all PCOS-related proteins.
2. How were PCOS-related proteins retrieved?
Below is the detailed flowchart on how PCOS-related proteins were compiled:
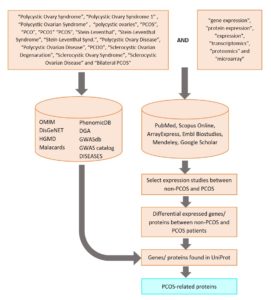
3. What are the differences between Databases and Resources datasets?
Both Databases and Resources are the datasets of the sources of PCOS-related proteins were collected from. Databases is the list of publicly available databases that were used to retrieve PCOS-related proteins. Meanwhile, Resources is the dataset of published expression studies (transcriptomics and proteomics) that were used to compile PCOS-related proteins.
4. What is the difference between interactor and partner in the Interactions dataset?
Interactor is a PCOS-related protein, while partner is a protein that is interacted with interactor (PCOS-related proteins).
5. Where is the score in Interactions dataset come from?
All of the scores of interactions were retrieved from HIPPIE database.
6. What is Publications dataset and how is it related to PCOS-related proteins?
Publications dataset is a list of all PCOS-related articles that were retrieved from PubMed. Based on PCOSBase schema, Publications table does not have a relationship with PCOS-related proteins table.
7. What is the disease vocabulary used for diseases?
All diseases in PCOSBase are mapped to the UMLS® CUIs. The UMLS® CUIs also are mapped to MesH ID to refer the class of the diseases.
8. What does it mean by ‘Expression’ column in the ‘Resource Details’ page?
Expression column is the pattern of expression of PCOS-related proteins in respective publication.
9. What does ‘Condition’ column in the ‘Resource Details’ page refer to?
Condition refers to the groups of biological samples for particular papers have. In Kenigsberg et al. 2009, they divide the samples into all non-PCOS vs. all PCOS, lean non-PCOS vs. lean PCOS, obese non-PCOS vs. obese PCOS, lean non-PCOS vs. obese non-PCOS and lean PCOS vs obese PCOS. Meanwhile, in Ouandaogo et al. 2012, they have isolated the samples in three different stages, which are germinal vesicle, metaphase I and metaphase II. Piltonen et al. 2o13 measured the gene expression of non-PCOS and PCOS women in four different tissues of endometrium, epithelial cells, endothelial cells, mesenchymal stem cells and stromal fibroblast.
10. How can I download the information in PCOSBase?
Every table in PCOSBase is provided with the “Download CSV” button at the top right of the table. Users can just click that button to download the particular data.
11. How to cite PCOSBase?
Nor Afiqah-Aleng, Sarahani Harun, Mohd Rusman Arief A-Rahman, Nor Azlan Nor Muhammad, Zeti-Azura Mohamed-Hussein; PCOSBase: a manually curated database of polycystic ovarian syndrome, Database, Volume 2017, 1 January 2017, bax098, https://doi.org/10.1093/database/bax098